Understanding genetic control of human protein N-glycosylation
Glycosylation – addition of carbohydrates – is a common cotranslational and posttranslational modification of proteins. Attachment of different glycans to the same protein can give rise to hundreds of its glycoforms. Different glycosylation of the same glycoprotein changes its physical properties as well as its biological function. In recent years, changes in different glycan abundances were associated with various diseases, including many of these we actively work on (e.g. chronic primary pain, see here). However, only a limited number of glycan structures are considered as established diagnostic biomarkers or therapeutic targets. This is in large part because interpretation of observed disease-associated glycan changes seen in various diseases is difficult due to limited knowledge about regulation of glycosylation: although biochemically, glycosylation pathway is well-studied, little is known about networks of genes that orchestrate cell- and tissues-specific regulation of these biochemical reactions in vivo, defining inter- and intra-individual differences.
Previously, we have implemented large studies to associate genome-wide polymorphisms with relative abundance of glycans harvested from specific protein, human immunoglobulin G (IgG)1,2. In recently published work3, led by prof. Gordan Lauc and PolyOmica’s Chief Scientist Yurii Aulchenko, we analysed glycans harvested form the totality of proteins found in human plasma. We discovered large number of new loci robustly associated with variation in plasma N-glycome, hypothesised the genes involved, and made inferences about the regulatory relations between them. This allowed us to demonstrate a clear overlap but also clear differences in genetic control between total plasma protein and IgG N-glycosylation.
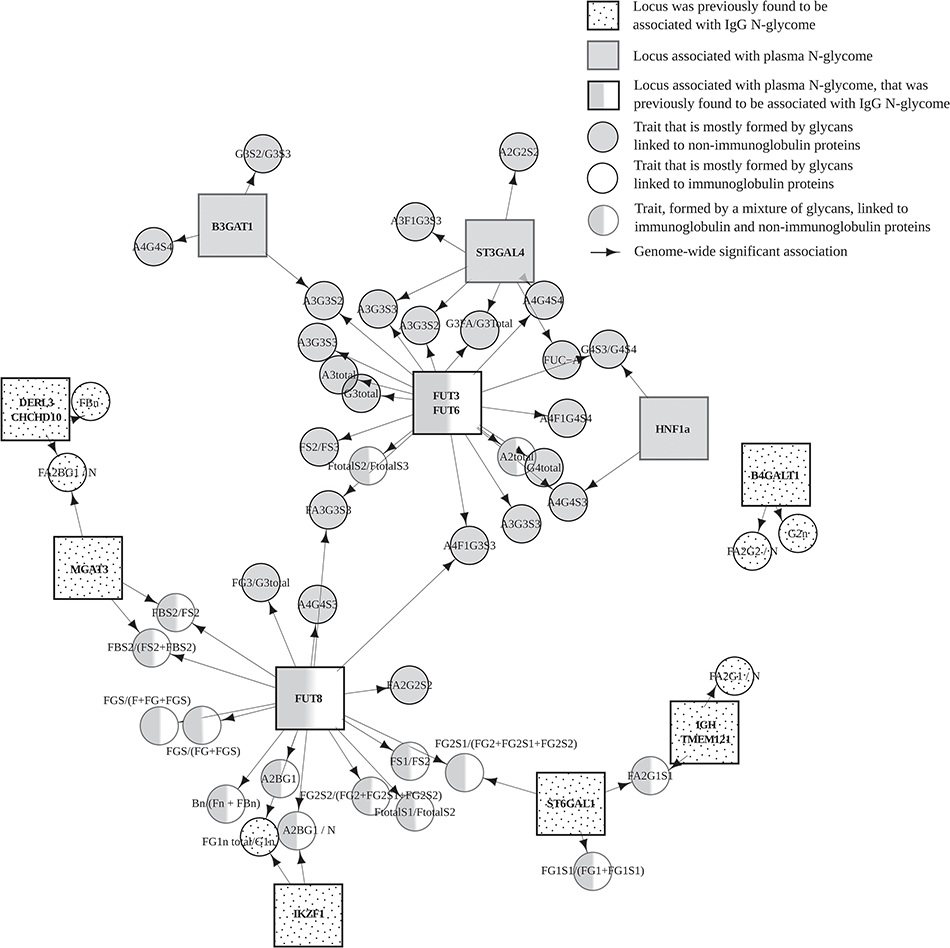
The majority of revealed loci contained genes that encode enzymes directly involved in glycosylation, but we also found loci that are possibly responsible for regulation of glycosylation process (e.g. DERL3, CHCHD10, TMEM121, IGH and IKZF1). Such regulatory genes, in our view, are plausible candidates that will help linking glycans with complex human diseases.
The figure shows a network view of associations between loci and glycan traits. Square nodes represent genetic loci labeled with the names of candidate gene(s), circle nodes represent glycan traits. Squares with polka dot pattern represent candidate genes, located in genomic regions that were previously found to be associated with IgG N-glycome. Grey squares represent candidate genes, located in genomic regions associated with plasma N-glycome. Grey/white squares represent candidate genes, located in genomic regions associated with plasma N-glycome, that were previously found to be associated with IgG N-glycome. Circles with polka dot pattern highlights glycan traits mostly containing glycans that are linked to immunoglobulins. Grey circles represent traits that are mostly formed by glycans linked to other (not immunoglobulin) proteins. Grey/white circles represent glycan traits, formed by a mixture of glycans that are linked to immunoglobulin and non-immunoglobulin proteins. Arrows represent genetic association (P < 1.66E−9) between gene and specific glycan.
- 1.Lauc G, Huffman JE, Pučić M, et al. Loci Associated with N-Glycosylation of Human Immunoglobulin G Show Pleiotropy with Autoimmune Diseases and Haematological Cancers. Gibson G, ed. PLoS Genet. January 2013:e1003225. doi:10.1371/journal.pgen.1003225
- 2.Shen X, Klarić L, Sharapov S, et al. Multivariate discovery and replication of five novel loci associated with Immunoglobulin G N-glycosylation. Nat Commun. September 2017. doi:10.1038/s41467-017-00453-3
- 3.Sharapov SZ, Tsepilov YA, Klaric L, et al. Defining the genetic control of human blood plasma N-glycome using genome-wide association study. Human Molecular Genetics. March 2019. doi:10.1093/hmg/ddz054
No Comments