Scientists from NSU and PolyKnomics developed one of the world largest databases of genetic associations
This database stores tens of billions of associations of genetic variants with human traits, investigated by the scientific community in hundreds of studies. Knowledge of such associations allows for better understanding of human genetics and biology and can contribute to the diagnosis, prevention and treatment of diseases. The results of the work were published in the journal “Nucleic Acids Research”.
Genome-wide association studies (GWAS) are the main tool for identifying genetic factors that affect the quantitative traits and risk of developing common human diseases. Knowledge of associations identified by GWAS helps to investigate the etiology of human diseases and to develop risk prediction models and can also be useful in the search for candidate biomarkers, therapeutic interventions and targets of such interventions. While the number of genetic associations studied by the scientific community is rapidly growing, the use of this data is hampered by their large volume and the lack of uniform standards for the format and quality.
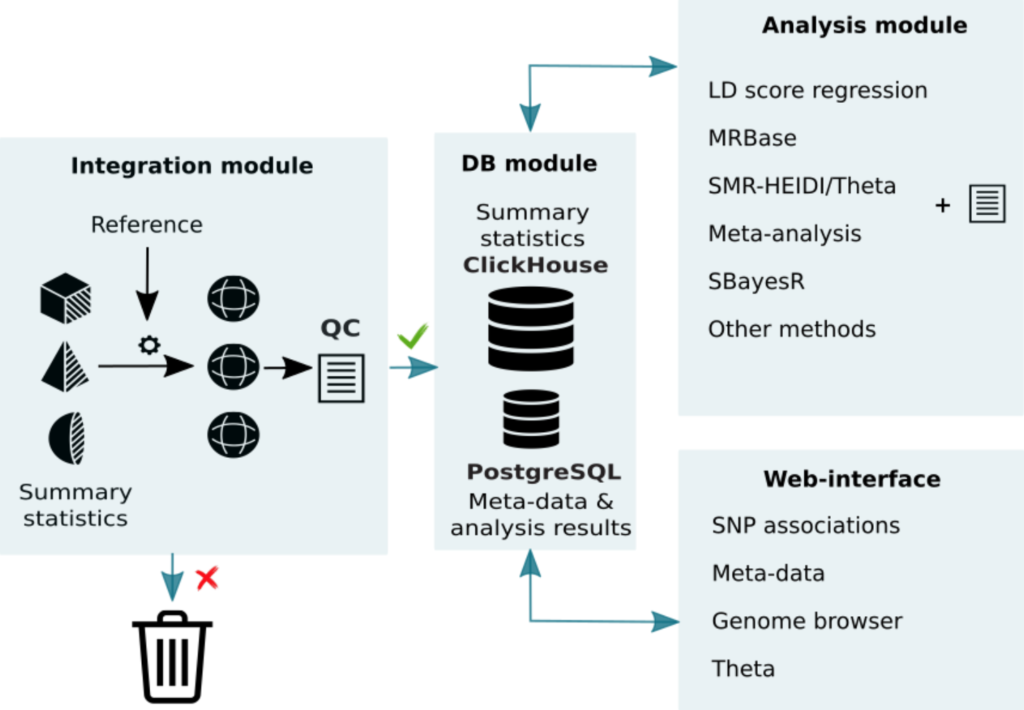
For many years, the scientists from the Laboratory of Theoretical and Applied Functional Genomics of the Novosibirsk State University (NSU, Russia), in collaboration with colleagues from PolyKnomics (the Netherlands), collected information on associations analysed in genetic studies, developed computational infrastructure and methods for unification, quality control and analysis of GWAS summary statistics. As a result of collecting and processing tens of terabytes of raw data, the researchers have obtained one of the world largest databases of genetic associations. The results were published in the Nucleic Acids Research journal1.
“We hope that the database of genetic associations we have developed will be useful for solving a wide range of problems: from fundamental studies of human genetics to the development of predictive models and search for candidate therapeutics,” — commented Tatyana Shashkova, researcher at the Laboratory of Theoretical and Applied Functional Genomics of the NSU.
The database contains the complete results of association studies of more than 7,000 traits, including quantitative traits, common diseases, and levels of metabolites, proteins and glycans, as well as the results of several large-scale studies of genetic control of gene expression. In total, the database contains data on more than 75 billion genetic associations. The PheLiGe web interface was developed to provide an access to the database. The team has also developed the GWAS-MAP platform2, which allows an access to the database and a wide range of analysis via the command line interface.
Lennart C. Karssen, CEO of PolyKnomics, adds: “The technological solution that we have developed together with NSU is multipurpose. For instance, it can be scaled to store and process information about millions of genomes. Such large data arrays emerge in the context of national biobanking programs or genomic breeding programs.”
- 1.Shashkova TI, Pakhomov ED, Gorev DD, Karssen LC, Joshi PK, Aulchenko YS. PheLiGe: an interactive database of billions of human genotype–phenotype associations. Nucleic Acids Research. Published online November 27, 2020:D1347-D1350. doi:10.1093/nar/gkaa1086
- 2.Shashkova TI, Gorev DD, Pakhomov ED, et al. The GWAS-MAP platform for aggregation of results of genome-wide association studies and the GWAS-MAP|homo database of 70 billion genetic associations of human traits. Vestn VOGiS. Published online December 31, 2020:876-884. doi:10.18699/vj20.686
No Comments