Identification of sixty three multigenic modules involved in susceptibility to inflammatory bowel disease
Inflammatory bowel disease (IBD) is a group of inflammatory conditions of the colon and small intestine. Crohn’s disease (CD) and ulcerative colitis (UC) are the principal types of inflammatory bowel disease. IBD was virtually unheard of in the early nineteen hundreds. Since then it’s incidence (as well as that of a series of other inflammatory diseases including asthma) has dramatically increased in industrialised societies: nowadays, more than one in 500 people will suffer from IBD at some point in their lifetime. This clearly points towards a key role for environmental and/or behavioural risk factors which, as of yet, haven’t been fully elucidated.
It appears, however, that we are not all equally sensitive to these risk factors, and that these inter-individual differences are to a large extent genetically determined. This has opened the possibility to use modern genetic approaches that have (recently) been shown to be very effective to decipher the molecular basis of these inter-individual differences as a means to gain new insights in the mechanisms underlying IBD as well as to identify novel drug targets.
Genome-wide association studies (GWAS) have identified more than 200 regions in the genome that underlie the inter-individual differences in the sensitivity to IBD1. However, before we can use genetic findings to better understand the disease and to discover new biomarker and intervention targets, we need to find the causative genes and hypothesise their relations to disease. Unfortunately, only for a handful of loci the causative functional variants and gene can be understood with confidence2, which is typical for complex diseases. Therefore we at PolyOmica concentrate on so-called “post-GWAS” studies, that allow linking the associated region to biological function.
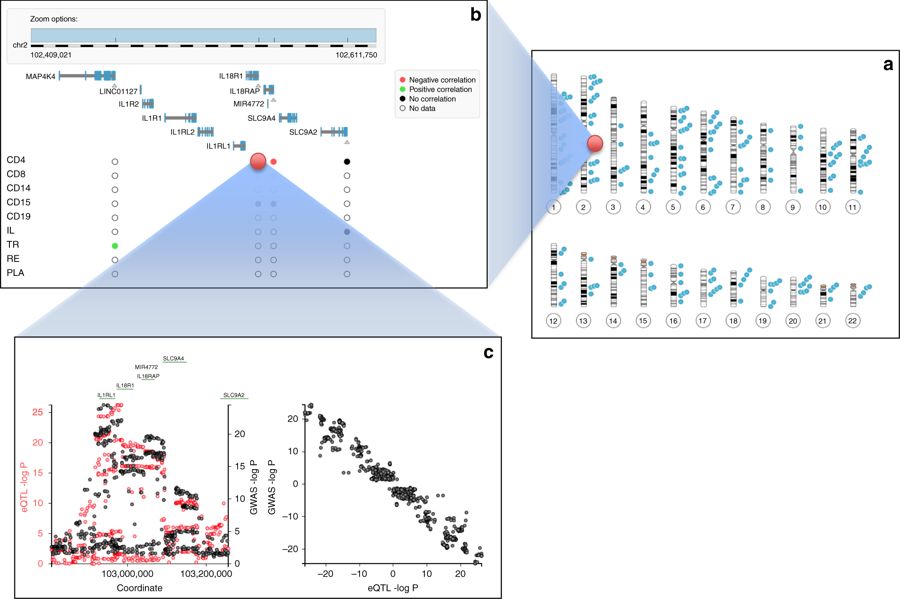
In a recent studies led by prof. Michel Georges, we have collaborated with researches from GIGA Institute at the University of Liège (ULg) and Novosibirsk State University (NSU) in a large-scale effort to identify genes targeted by the variants associated with risk of IBD. We have searched for genes whose expression level (rather than the amino-acid sequence) is perturbed in susceptible individuals using an analysis of cis-eQTL. A unique dataset, CEDAR, involving nine disease-relevant cell types in more than 300 individuals was generated, and the expression of close to 2,000 genes mapping to the 200 IBD risk loci have been studied. By applying newly developed methods, we have found 99 genes in 63 of the 200 IBD risk loci that are very likely to be directly implicated in triggering IBD. These constitute a very rich source of potential new targets for drug development. In order to to share the ensuing results with the scientific community, we have developed an open-access CEDAR website that visualises correlated disease and expression association patterns within their genomic context. The image at the top of the page presents screenshots of the CEDAR website, showing a) known Crohn’s Disease risk loci on the human karyotype, b) a zoom in the HD35 risk locus showing the gene content and summarizing local CEDAR cis-eQTL data, and c) a zoom in the association pattern for Crohn’s disease (black) and expression for IL18R1 (red), as well as the signed correlation between these association patterns.
The results have been published in the prestigious scientific journal Nature Communications3. The press-release of University of Liege was published on June 21, 2018.
No Comments