database, genomics, GWAS, software
Scientists from NSU and PolyKnomics developed one of the world largest databases of genetic associations
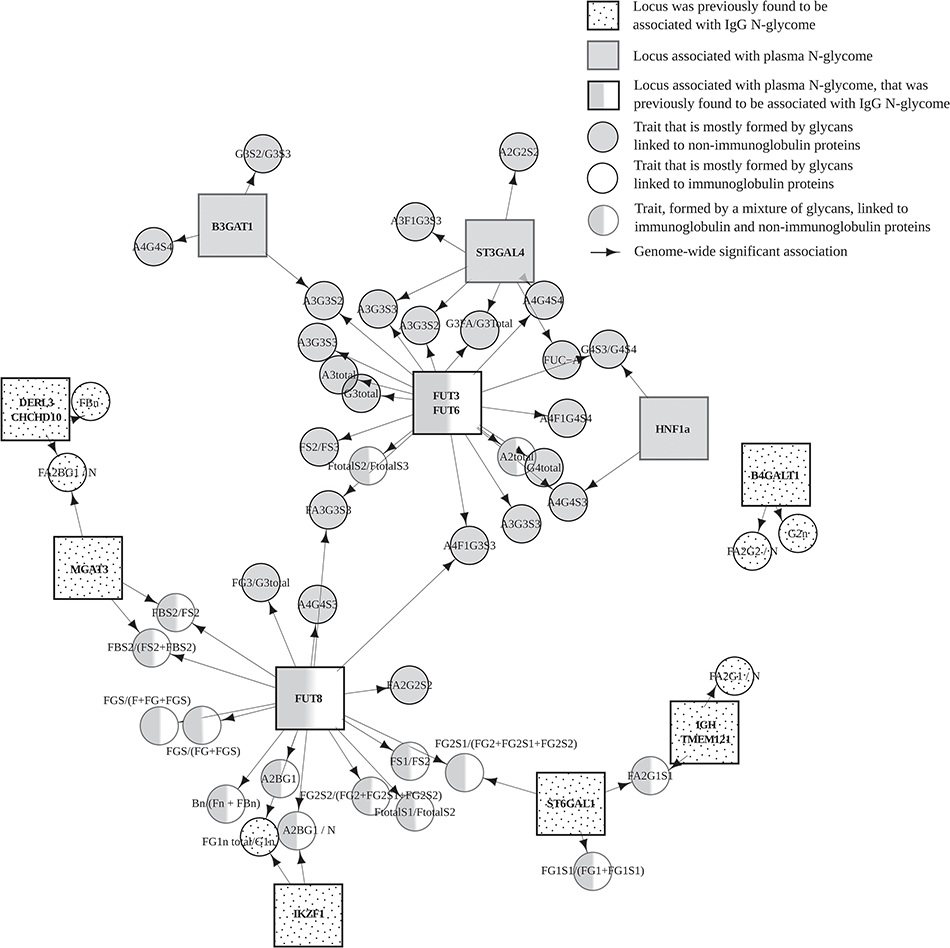
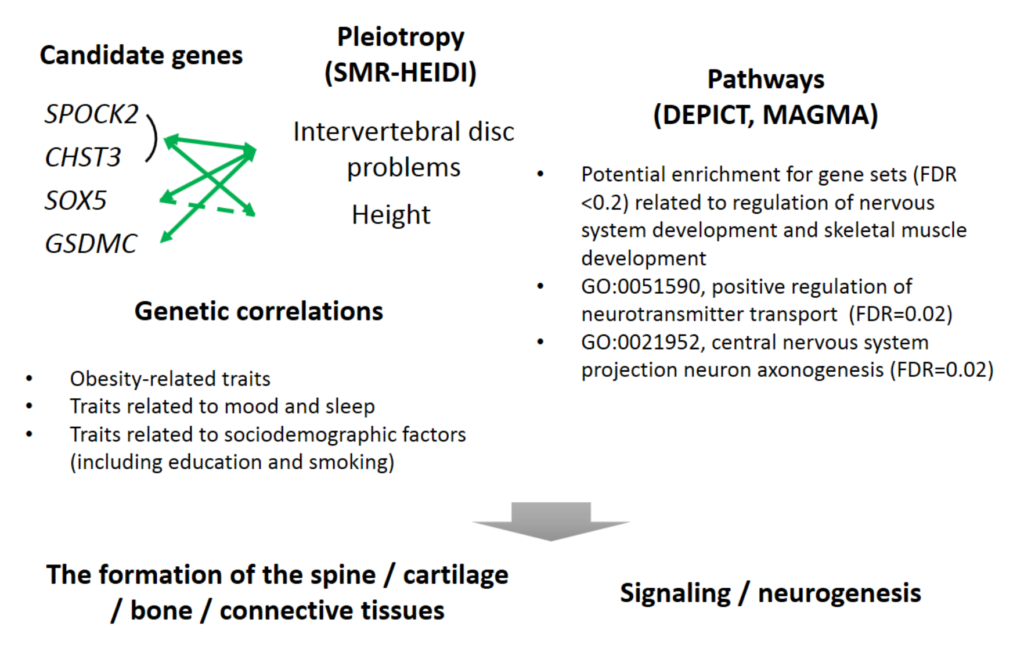
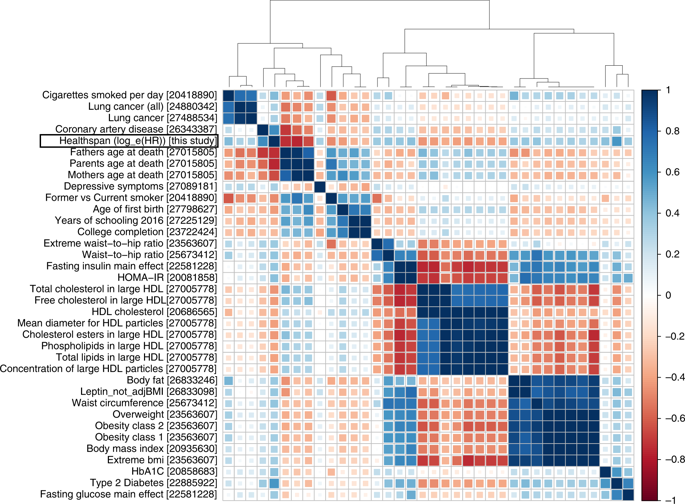
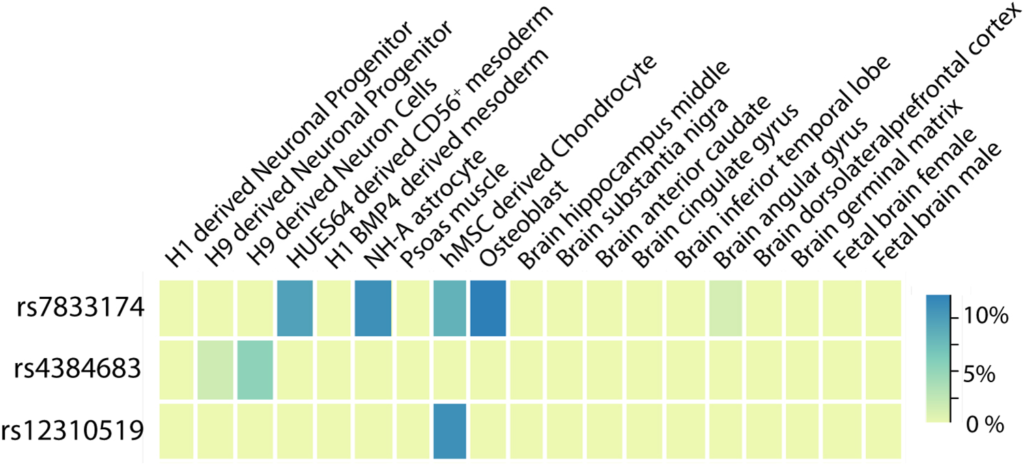
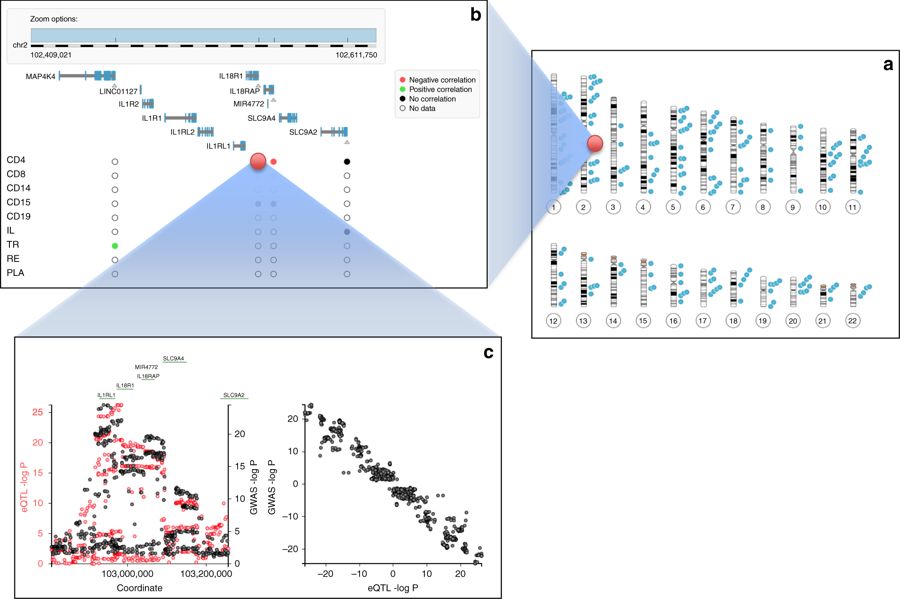
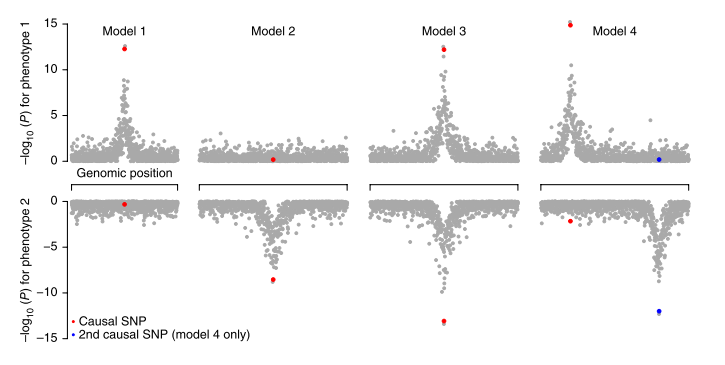
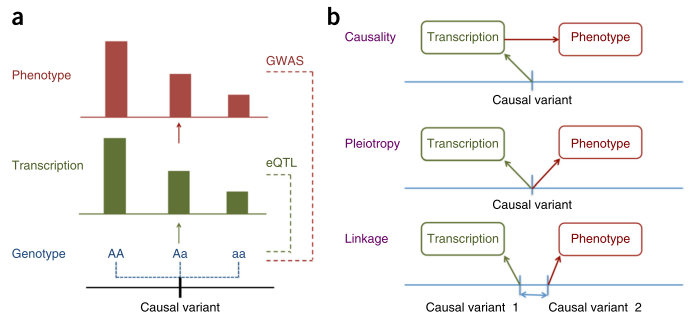
This database stores tens of billions of associations of genetic variants with human traits, investigated by the scientific community in hundreds of studies. Knowledge of such associations allows for better understanding of human genetics and biology and can contribute to the diagnosis, prevention and treatment of diseases. The results of the work were published in […]
Read full case study