
In a little more than two weeks from now I will be at the ErasmusMC university medical centre to teach the “Linux for Scientists” course. The EL016 course, as it is officially called, consists of two full days of teaching and practice and is scheduled on January 31st and February 1st, 2023. It is part […]
Continue reading
Posted on 13 January 2023 by Lennart Karssen

Time flies! Since the end of July I have been working not from the Netherlands as usual, but from Uganda. Now, this “expat period” is coming to an end and my family and I will be back in Den Bosch in January. All in all, I think that my working remotely has not affected the […]
Continue reading

In two weeks’ time, on Wednesday 26th and Thursday 27th of January, I will be two full days at the Erasmus Medical Centre, Rotterdam. I will be teaching the course Linux for Scientists for students of the Netherlands Institute for Health Sciences (NIHES) MSc programmes.
Continue reading
This database stores tens of billions of associations of genetic variants with human traits, investigated by the scientific community in hundreds of studies. Knowledge of such associations allows for better understanding of human genetics and biology and can contribute to the diagnosis, prevention and treatment of diseases. The results of the work were published in […]
Continue reading
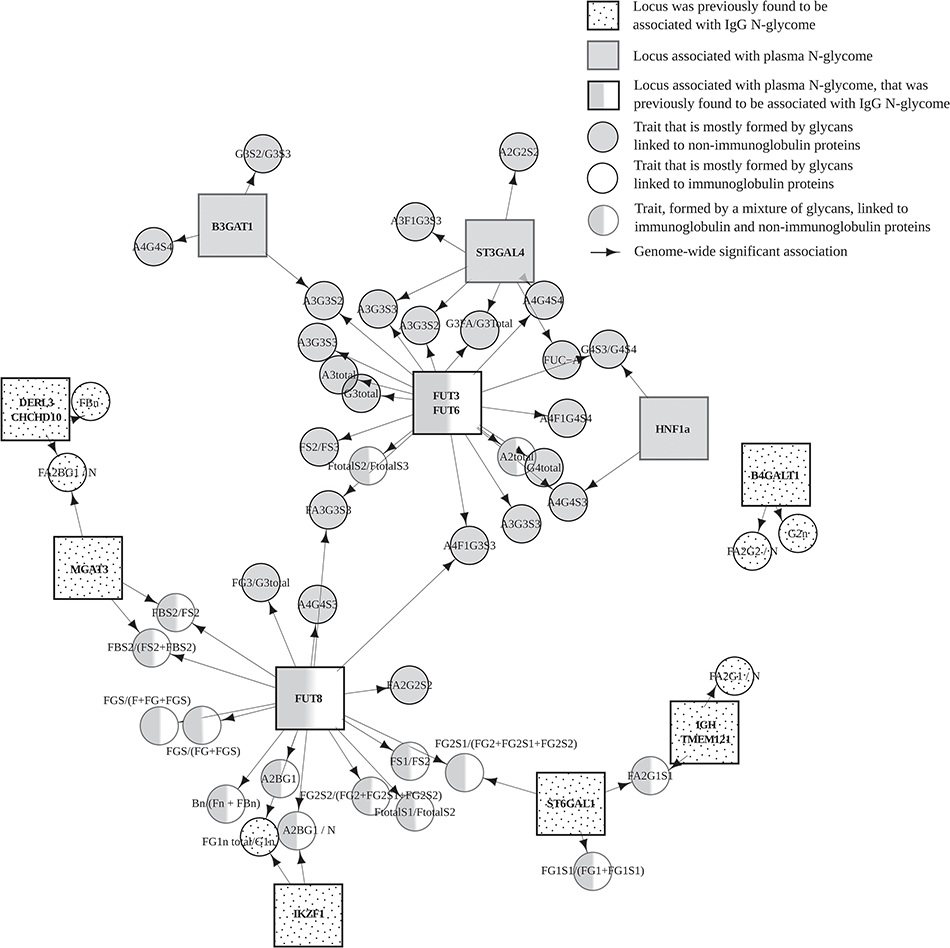
Immunoglobulin G (IgG) that plays central role in adaptive immunity is the most abundant immunoglobulin found in human plasma. All IgG molecules are N-glycosylated. N-glycosylation of IgG contributes to the maintenance of its structure, stability, and solubility. In a large collaborative study, we analysed association between genomes and glycosylation of immunoglobulin G (IgG) in more than […]
Continue reading
A team of six, with two authors affiliated with PolyOmica, has received a 2020 prize in Clinical Science from the International Society for the Study of the Lumbar Spine (ISSLS) for a study that found a significant causal effect of BMI on both back pain and chronic back pain1. Back pain is the #1 cause […]
Continue reading
Glycosylation – addition of carbohydrates – is a common cotranslational and posttranslational modification of proteins. Attachment of different glycans to the same protein can give rise to hundreds of its glycoforms. Different glycosylation of the same glycoprotein changes its physical properties as well as its biological function. In recent years, changes in different glycan abundances were associated with various diseases, including many of these […]
Continue reading
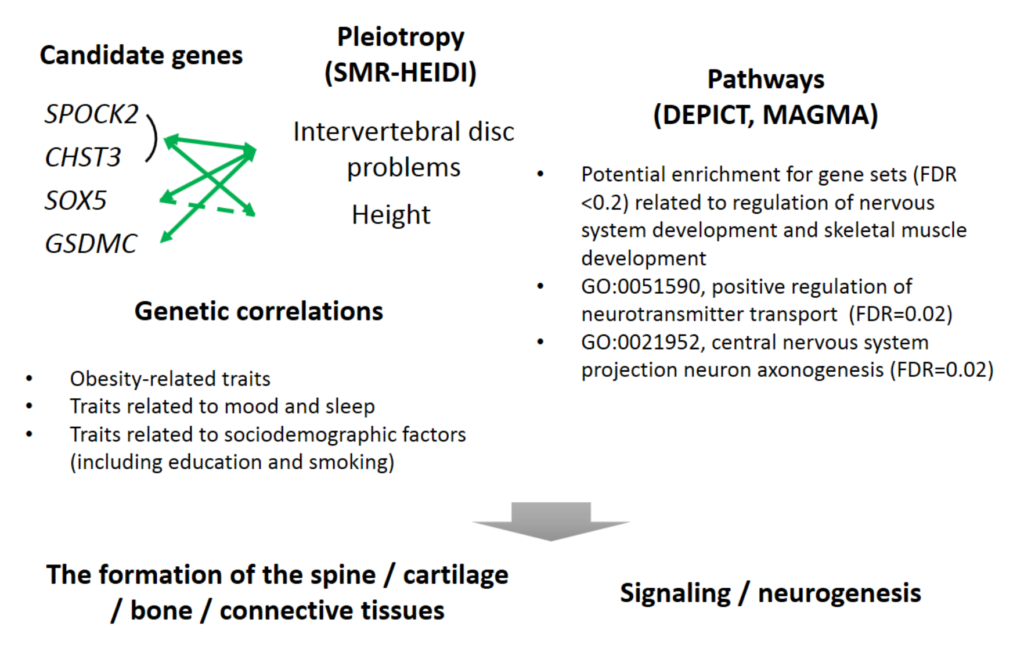
Back pain is the number one cause of years lived with disability worldwide and one of the most common reasons for health care visits in developed countries, yet surprisingly little is known regarding the biology underlying this symptom. In our previous study1, published in Sept 2018, we identified three novel genetic variants associated with chronic […]
Continue reading
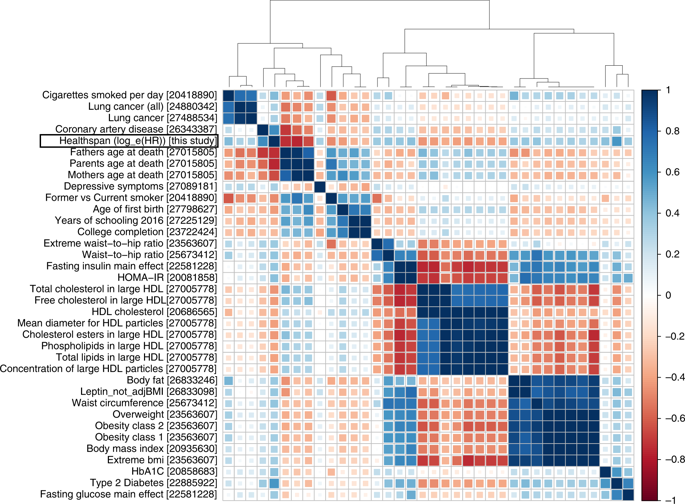
Aging populations face diminishing quality of life due to increased disease and morbidity. These challenges call for longevity research to focus on understanding the pathways controlling healthspan. A new study, led by dr. Peter Fedichev (Gero LLC) and prof. Yurii Aulchenko (PolyOmica), used the data from the UK Biobank to understand biology of healthspan1. We observed that the […]
Continue reading
Posted on 12 December 2018 by Lennart Karssen
The Clarivate Analytics list of Highly Cited Researchers for 2018 identifies scientists who have demonstrated significant influence through the publication of multiple highly cited papers in the last decade. PolyOmica’s Chief Scientist, Yurii Aulchenko , was selected for his substantial influence across several fields in the period 2006–2016. Approximately 6,000 researchers are named Highly Cited Researchers in […]
Continue reading
Adding covariates can either increase or decrease the power of finding an effect of a predictor on the outcome of interest. In a recent publication in GigaScience we used knowledge of biochemical networks to select covariates for conditional genetic association analysis. We demonstrated that the power depends on whether genes and environment play along or […]
Continue reading
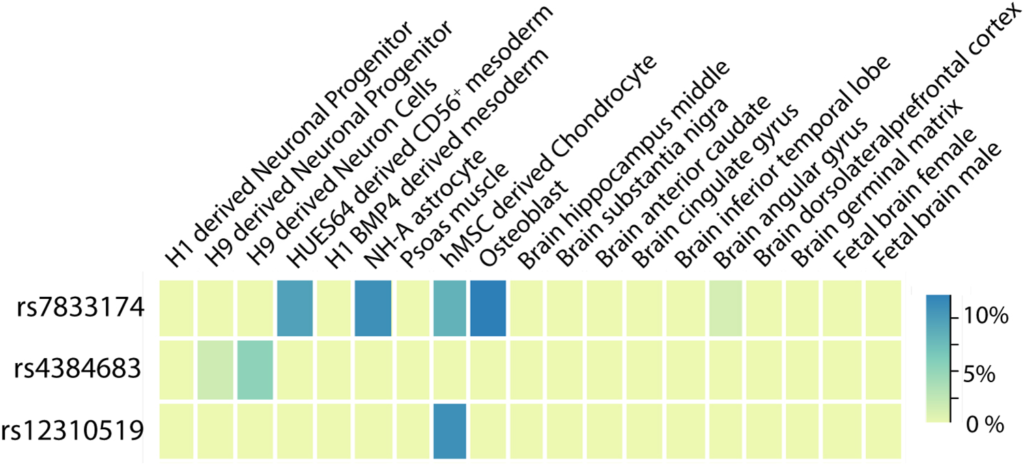
Back pain is the #1 cause of years lived with disability worldwide and one of the most common reasons for health care visits in developed countries, yet surprisingly little is known regarding the biology underlying this symptom. A new study, led by Pradeep Suri of the Department of Veterans Affairs in Seattle, Washington (U.S.A.) and […]
Continue reading
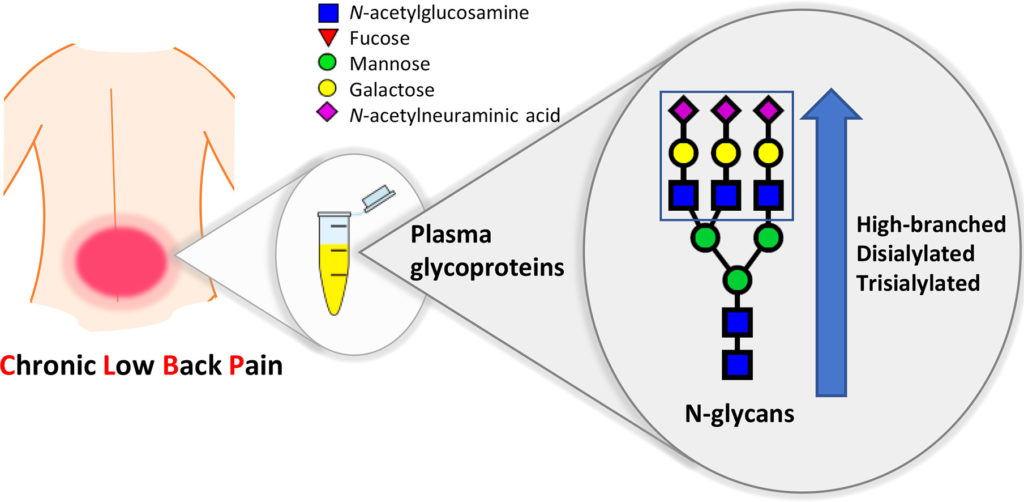
Last Thursday, July 5, 2018, a manuscript1 reporting results from a large multicentric case-control study of patients with chronic lower back pain (CLBP) was published in Biochimica et Biophysica Acta (BBA) – General Subjects. The work was funded by the EU FP7 project PainOmics in which we participate. The study of plasma N-glycome composition in patients […]
Continue reading
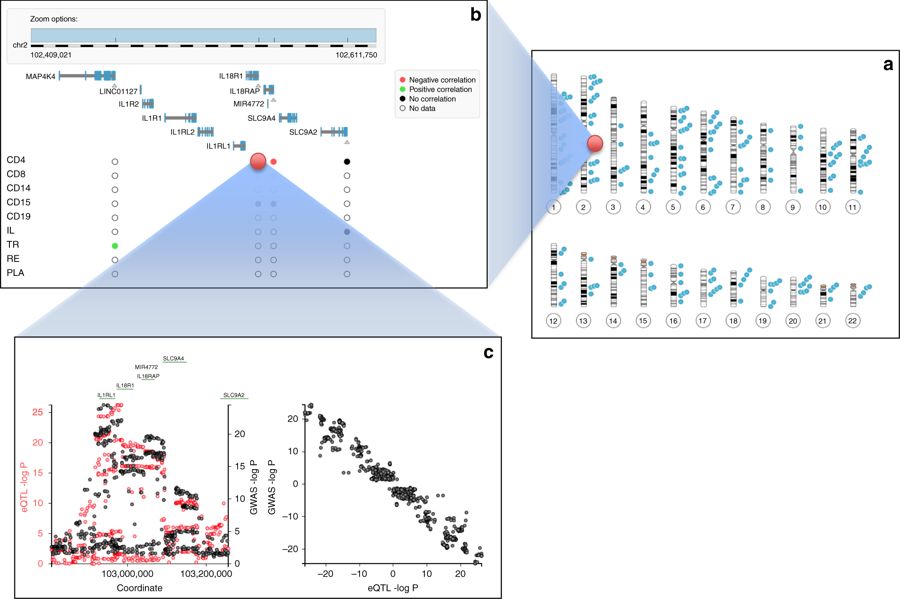
Inflammatory bowel disease (IBD) is a group of inflammatory conditions of the colon and small intestine. Crohn’s disease (CD) and ulcerative colitis (UC) are the principal types of inflammatory bowel disease. IBD was virtually unheard of in the early nineteen hundreds. Since then it’s incidence (as well as that of a series of other inflammatory diseases including asthma) has dramatically […]
Continue reading

From March 26–28, 2018 the IMforFUTURE International Training Network (ITN), in which PolyOmica is a partner, will have its first network meeting. The meeting will be held at the BioCentar in Zagreb, Croatia, and aside from various committee meetings and social events, the meeting will consist of training sessions for the fellows enrolled in the […]
Continue reading

On Monday March 5 2018, I will be teaching at the NIHES course “An Introduction to the Analysis of Next-Generation Sequencing data” at the Erasmus Medical Centre in Rotterdam. In this one-week course students will learn the basics of how NGS data is generated and how to analyse this type of data. PolyOmica’s contribution to […]
Continue reading

In PolyOmica, we some times need to train people in the basics of genetics and genomics. While training people in basics is fun, it is quite time consuming, and is not exactly our company’s core activity. For a long time, we thought that it would be great to have a massive open online course (MOOC) […]
Continue reading

Back in 2014 we initiated the Research Summer School in Statistical Omics with the help of many friends and colleagues, and with the support (financial and other) from various sources, including the EU-funded MIMOmics project, in which we participate, and the EU-funded Integra-Life project. In a period of two weeks we immersed 13 students (and […]
Continue reading

From 20 till 27 August, the EU FP7 consortium MIMOmics in which PolyOmica is a partner, organises a Summer School including a Workshop in Omics Studies. The goal of the School is to train a new generation of statistical omics scientists with a biological and biotechnological background to enable them to understand and to apply […]
Continue reading
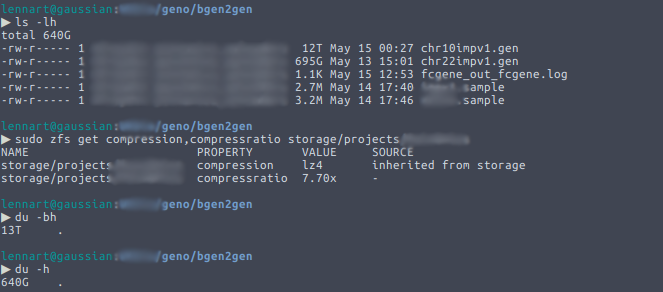
One of the great things of ZFS is its built-in compression capability. Enabling compression on a ZFS filesystem allows you to save a significant amount of space, especially in a field like ours where one sometimes needs to use large plain text files. It is completely transparent: the user doesn’t notice anything. Well… unless they […]
Continue reading

Next week (starting Monday January 30 2017) Yurii and I will be in Rotterdam at the Erasmus University Medical Center where we will teach in the “Advances in Genome-Wide Associations” course of NIHES. This course is part of the Master’s programme in Health Sciences and is compulsory for those who follow the specialisation “Genetic Epidemiology”. […]
Continue reading
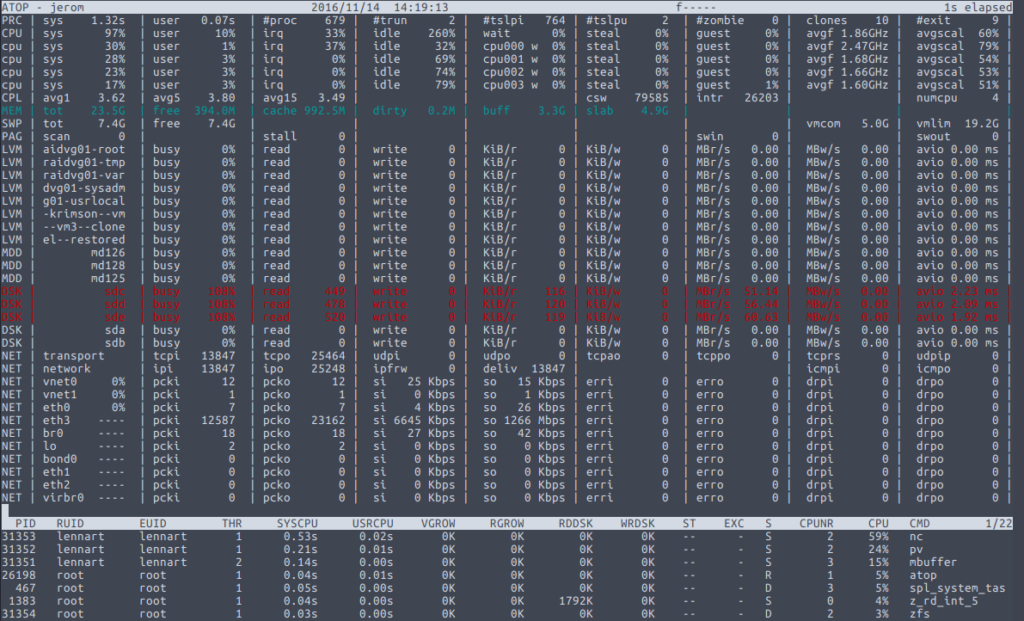
I recently had to send a couple of ZFS file systems from one server to another, both running Ubuntu Linux 16.04. Since both servers were in the same trusted local network, there was no need for encryption. As various other websites will tell you, the fastest way to transfer data over a local network is […]
Continue reading
We are currently preparing for a visit to Zagreb, Croatia, where we will attend the fourth annual meeting of the MIMOmics EU project on which we collaborate. The programme will covers topics like glycomics and metabolomics, statistical prediction based on multiple omics, casual inference, networks and epidemiology. So if you are in the vicinity of […]
Continue reading
coffee, eQTL, GWAS, paper Posted on 29 August 2016 by Lennart Karssen

Sometimes one gets unexpected news. Like I got a couple of days ago when one of our collaborators, Nicola Pirastu, informed me that the press release of one of our recent papers was apparently picked up by the ‘regular’ press. At the time of writing a search on news.google.com gives four pages of returned results […]
Continue reading

We have just returned from Split, Croatia, where we organised and co-sponsored the third Research School in Statistical Omics (RSSSO2016). With this Summer School we aim to train young researchers (at bachelor, master or PhD level) with a bio{logical,medical,chemical} background and interest in quantitative statistical methods to become the next generation of life scientists; these […]
Continue reading

Recently we published an overview paper of the GenABEL Project [1] on F1000Research. The GenABEL Project is a framework for collaborative, robust, transparent, open-source based development of statistical genomics methodology that PolyOmica sponsors. One of the reasons for publishing this overview paper was that we felt the need for a single, easy-to-point-to reference for the […]
Continue reading
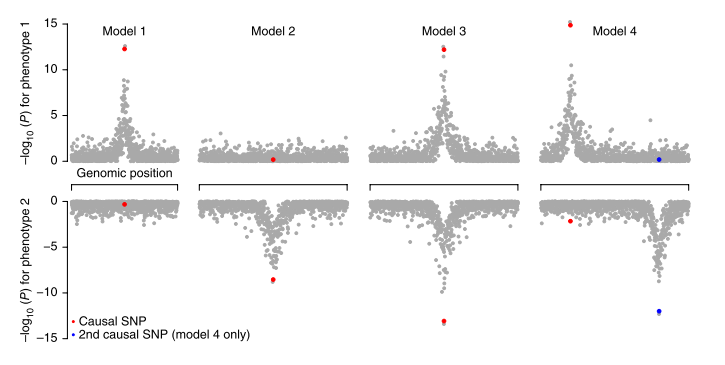
Pickrell et al. (Pickrell et al., 2016) come up with a very nice approach which uses summary-level data to detect variants exhibiting effects on multiple traits. They also propose use of multi-locus information to infer causative relations between traits. The manuscript starts with reminding the reader that the observed association between a genetic variant and multiple […]
Continue reading
One of the research projects we are working on at PolyOmica is the EU FP7-funded project MIMOmics (Methods for Integrated analysis of multiple Omics datasets). Last week, the partners in work package 6 (WP6) “Data integration and distributing computing” met in Helsinki for a two-day hackathon. The goal of the hackathon was to work on […]
Continue reading
This is a follow-up post to the mini-review of the work of Zhu et al. [1] and is a reaction to tweet from @JosephPowell_UQ 🙂 In short, in Zhu et al. [1] have used eQTL results from peripheral blood to answer the question of potential biological function affected by genetic variation associated with five complex trait. Using such eQTLs may […]
Continue reading
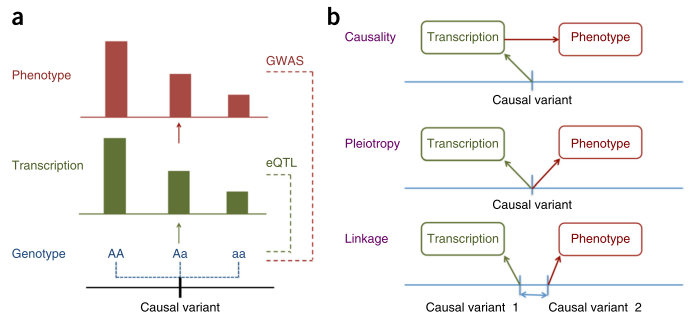
In this work, Zhu and colleagues [1] start with asking the question whether it would be possible to use summary-level data coming from GWAS of a complex trait coupled with eQTL association results to address the question of causality. Could the association observed for the complex trait be explained by changes in the level of a […]
Continue reading
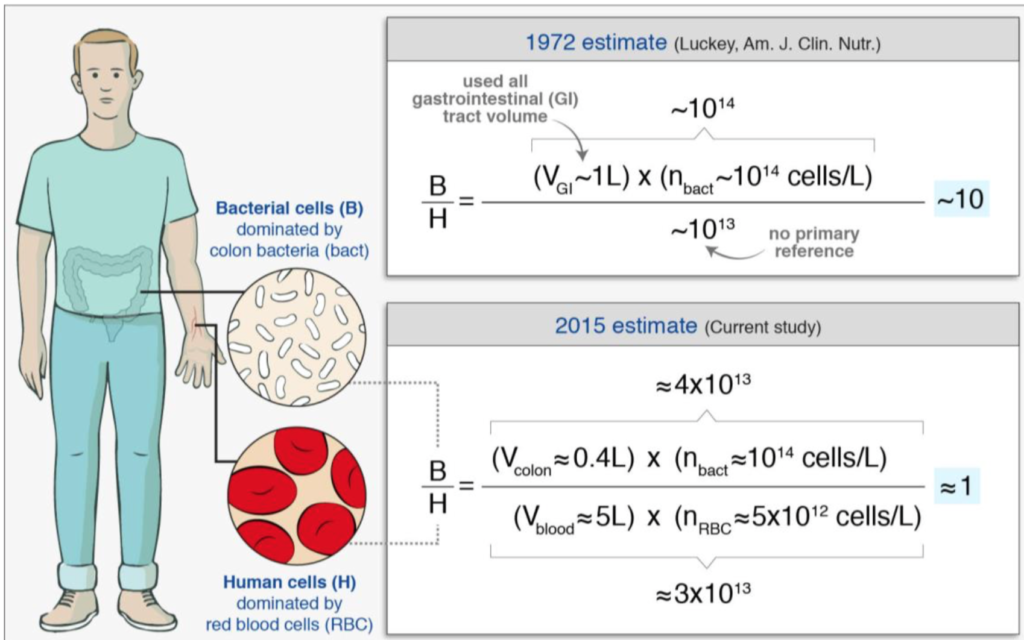
With interest we read a review/analysis paper [1] which questions the ‘common knowledge’ that we have ten times more bacteria on/in us than we we have cells. The authors make an interesting historical analysis tracing the ’10:1′ statement back to its origins (back to the beginning of 1970s!); based on analysis of more recent literature, they […]
Continue reading
Posted on 30 May 2016 by Lennart Karssen
Dear visitors of the PolyOmica blog. This is the first post on our new website. We are very excited about having this new platform at disposal and plan to inform you regularly of the things we are working on. On this blog you can expect announcements of things we do, like upcoming courses, but also […]
Continue reading



















